
Plant Genetic Engineering Laboratory
Our Research
Genome Editing for Crop Improvement




The advent of CRISPR-based genome editing is revolutionizing plant biology and crop improvement. The new technology promises to deliver more nutritious food and better crops with higher yields, resistant to diseases and better adapted to climate change. CRISPR-based breeding can be achieved in record times with considerable savings. Nevertheless, as any new technology, there are still bottlenecks and problems that need to be solved to reach its full potential. We are working on the development of new CRISPR platform technologies to speed up and facilitate the application of CRISPR to elite crop varieties. We are mostly focusing on transgenic-free approaches able to introduce genome edits without the need to produce a first GM generation of plants.

Plant Disease Signaling

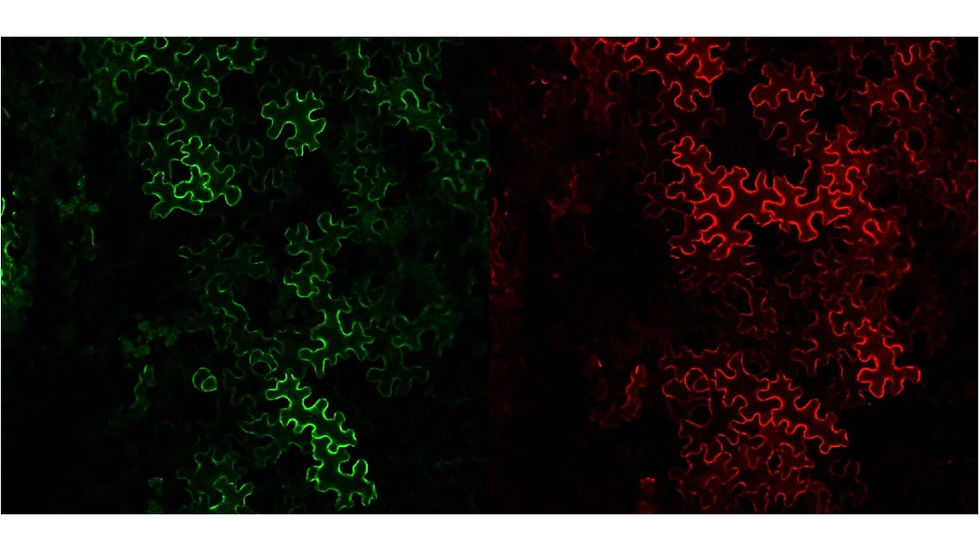

Plants are exposed to virtually thousands of enemies with extremely different lifestyles and strategies to invade their hosts. Surprisingly, for each plant species, only a few pathogens are successful thanks to very sophisticated defence mechanisms that they have evolved over thousands of years. There are many ways in which plants defend themselves against pathogens and most of them start the same way: the plant needs to know that it is under attack. Early sensing of a pathogen is essential to deploy an adequate response and ensure survival. Once a plant cell has determined that it is being attacked it needs to communicate with its nucleus to activate defence genes and other cells to give them early warning. We are studying such communications at all levels; from the receptors at the cell surface to the intermediates carrying the signal and finally the cellular response that allows the demise of the pathogen. We have discovered a new defence component called heterotrimeric G-proteins that play a very important role in the plant immunity. Our research is trying to discover how it works at the molecular and cellular levels.

Plant Signal Transduction


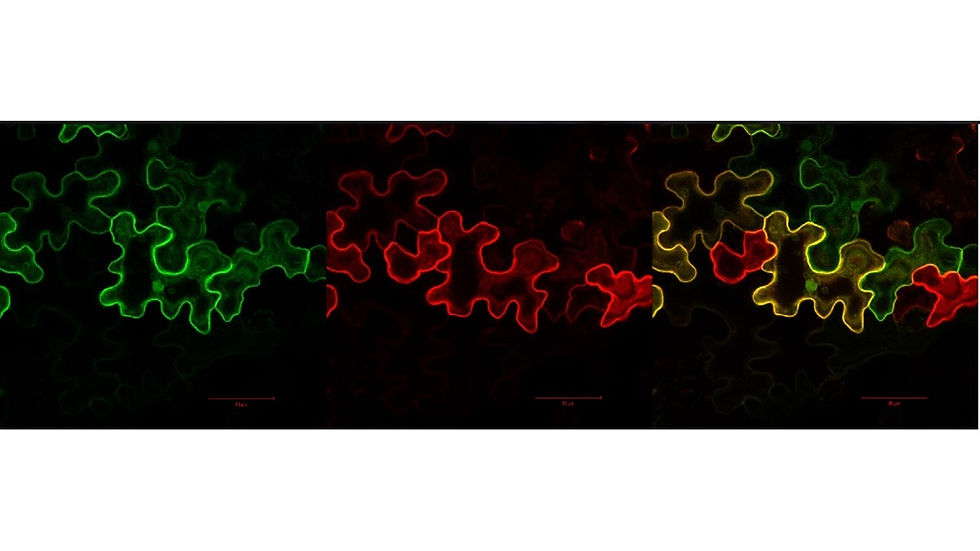
Plants need to perceive and accordingly respond to an incredible number of signals from their surrounding environment in order to survive. Aside from the sheer number of them, the nature of these signals can be extremely variable; chemical nutrients in the rhizosphere, day length (or more correctly, night), pathogenic microbes, water shortage, heat, cold, herbivores, gravity, etc. In order to recognize such a varied array of signals they have evolved a large number of receptors with very different structural characteristics. However, perception of a stimulus is only the first step in a long and complicated process culminating in the production of correct responses that can be as diverse as the original stimuli. Coupling each specific stimulus to the appropriate response is essential and can be the difference between life and death. Aside from specificity, controlling the magnitude of the response is also important and needs to be proportionate to the perceived signal. Finally, while some responses such as flowering in response to photoperiod can be produced over a relatively wide window of time, in other cases speed is of the essence and stomatas need to be quickly closed in a hot mid-day before dehydration results in irreversible damage to the plant. In order to control the specificity, speed and magnitude of the response, plants have evolved sophisticated signal transduction mechanisms that in most cases are far from linear events. A single stimulus can trigger one or more receptors and each activated receptor can initiate several signalling cascades. To complicate it all, signalling to different stimuli can have extensive cross-talk as is the case in the response to hormones.
We are studying a family of signal transduction molecules called heterotrimeric G-proteins (G proteins) consisting of α, β and γ subunits that mediate many important signalling pathways in eukaryotes. They are involved in morphological development cell proliferation, ion-channel regulation, stomatal control, light perception, early seedling development, abiotic stresses and, most importantly, yield in very important crops such as rice.
Classically, ligand bound 7-transmembrane (7TM) spanning G protein-coupled receptors (GPCRs) catalyse the exchange of bound GDP for GTP on Gα, resulting in activation of the heterotrimer and dissociation of the two functional elements, the Gα subunit and the Gβγ dimer. Gα and Gβγ independently interact with multiple downstream effectors mediating specific signal transduction pathways. Eventually, the intrinsic GTPase activity of Gα hydrolyses GTP to GDP increasing its affinity for the Gβγ dimer, leading to the re-association of the inactive heterotrimer at the receptor.
Our group has made important contributions to the available knowledge of plant G proteins discovered several characteristics that make plant G proteins very different from their animal counterparts. For example we reported the first plant Gγ subunit (Mason and Botella 2001) and recently discovered a new class of Gγ proteins never before described in plant or animal systems, proving their involvement in development and guard cell physiology (Chakravorty et al. 2011). We also reported that two of the most important QTLs for rice yield are members of the new Gγ subunit family. We then provided a mechanistic model to explain the apparently contradictory results obtained by a number of groups working on these QTLs (Botella 2012). In our last published contribution, we have identified a set of completely new subunits to the heterotrimer. We have proven that, contrary to the current belief, XLG proteins can indeed be part of the G protein heterotrimer and replace the Gα subunit to form functional associations with the Gβγ dimer (Maruta et al. 2015).
Our research is now focused on establishing the molecular signalling relay used by G proteins to control all the above mentioned processes.

Disease diagnostics for rich and poor countries

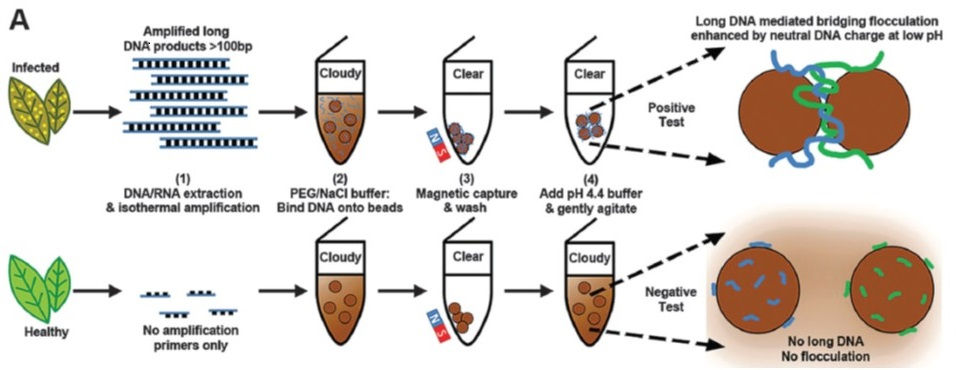


Agriculture is a major economic activity with a total annual value of $1500 billion US dollars. However, up to a third of the agricultural production is lost due to three major causes: disease outbreaks, insect attack and weed competition. Among them, losses caused by crop diseases are the most important issue globally, especially in agriculturally reliant countries. In the absence of resistance, the ideal method to control disease outbreaks is by early detection in the field before it spreads to neighbouring farms. On-site, quick and cheap pathogen detection is the holy grail of disease diagnostics.
We have developed a number of technologies to detect crop pathogens using different combinations of molecular biology and nanotechnology. One of them uses Molecular Inversion Probes and can be multiplexed to detect several pathogens simultaneously. Our latest development is called Single-Drop Genomics (SDG): a comprehensive, field-ready assay for on-site pathogen detection. SDG combines a simple yet accurate on-site sampling process with a robust isothermal amplification and a novel naked-eye evaluation providing Yes/No results. SDG has been successfully used to detect fungal and bacterial pathogens and RNA-based viruses in diseased plants, including commercial bananas. SDG also detected pathogens in livestock and environmental samples. Finally, SDG’s universality was established by detecting causal pathogens for human diseases including HIV, tuberculosis, malaria and influenza. SDG does not need any electrical power supply, can be supplied as a kit without refrigeration, performed in the field by minimally trained personnel and yielding results in under 90 minutes at very low cost (~$2 USD).

Biotechnological control of crop diseases



The best way to control diseases and benefit farmers and the environment is to produce genetically superior crops that can protect themselves against pathogens without the use of chemicals. Biotechnology and genetic engineering have open the door for new and more efficient approaches to disease control. In our laboratory we are discovering new genes with biotechnological applications as well as developing new platform technologies that provide resistance against many diseases and can be applied to any existing crop. We developed the Host-delivered RNAi (HD-RNAi) as an effective strategy to silence genes in plant parasitic nematodes and have now extended its use to fungal pathogens. New strategies have now been developed that have resulted in a 100% resistance to the devastating pathogen Fusarium, which cause large losses in many of the most important crops, including bananas and wheat.